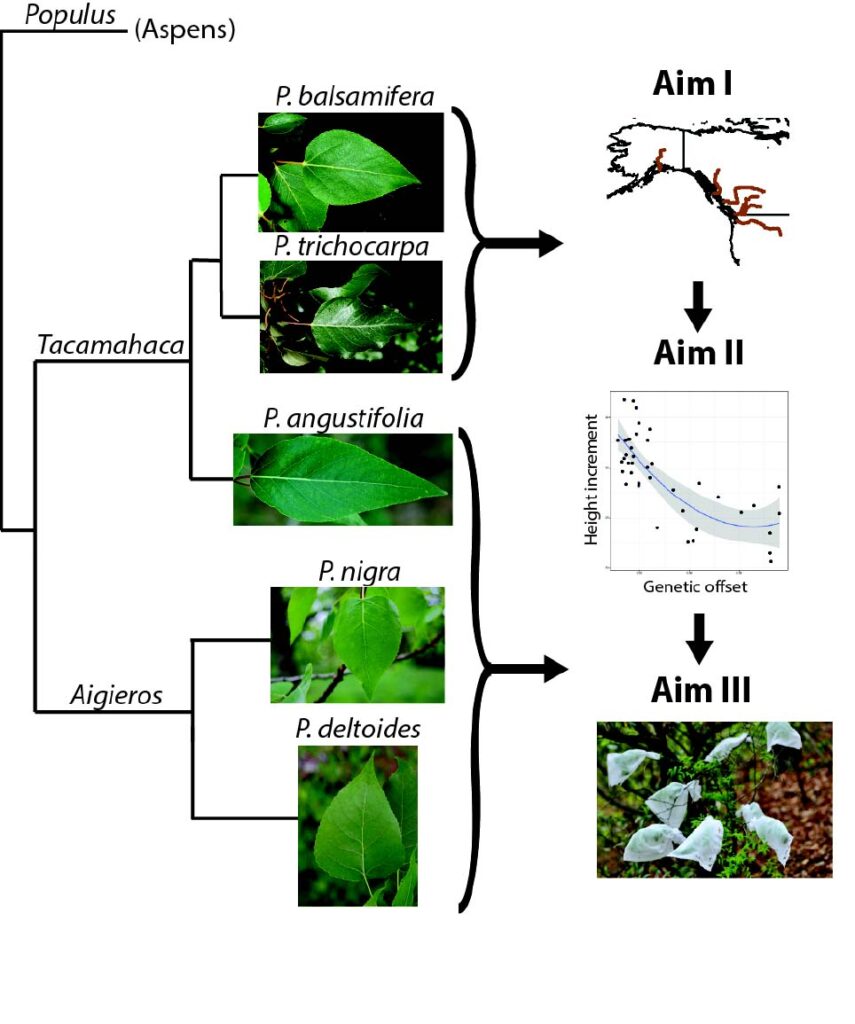
Adaptation to Climate In Temperate and Boreal Forest Trees
We use a variety of genomic and statistical tools to dissect the genetic architecture of complex traits, and to understand how the constraints imposed by population history may impinge on future adaptive evolution. Through an NSF CAREER award, we generated genome-wide re-sequencing data to understand the genomic basis for adaptation to climate in Populus trichocarpa. We used exome capture to sequence the gene space of a mapping population comprised of 500 poplar clones spanning the latitudinal and elevational range of the species. With these data, we demonstrated the utility of sequence capture as a tool for forest genomic research (Zhou & Holliday 2012), showed a role for regulatory sites in local adaptation (Zhou et al. 2014), showed that the genomic architecture of adaptation depends on the magnitude of gene flow (Holliday et al. 2016), demonstrated that deleterious alleles impinge on the adaptive capacity of poplar (Zhang et al. 2016), and used genome-wide association mapping to show that parallel altitudinal and latitudinal clines can be partially explained by parallelism at the genomic level (Zhang et al. 2019).


Genomics to Accelerate American Chestnut Restoration
American chestnut (Castanea dentata) was once the most economically and ecologically important hardwood species in the United States. In the first half of the 20th century, an exotic fungal pathogen – Cryphonectria parasitica – decimated the species, killing approximately four billion trees. The American Chestnut Foundation (TACF) has used hybrid breeding to introgress blight resistance alleles from Chinese chestnut (Castanea mollissima) into American chestnut, and we recently completed a NIFA-funded project to develop an accurate genomic selection model to accelerate the selection process in TACF seed orchards. TACF and collaborators have also developed American chestnut lines expressing a wheat oxalate oxidase (OxO) transgene that show levels of resistance comparable to that of Chinese chestnut. We were awarded a NIFA Foundational grant to study local adaptation in remnant chestnut re-sprouts, with the goal of crossing the transgenic founder lines with wild germplasm to produce diverse, locally adapted reforestation populations.

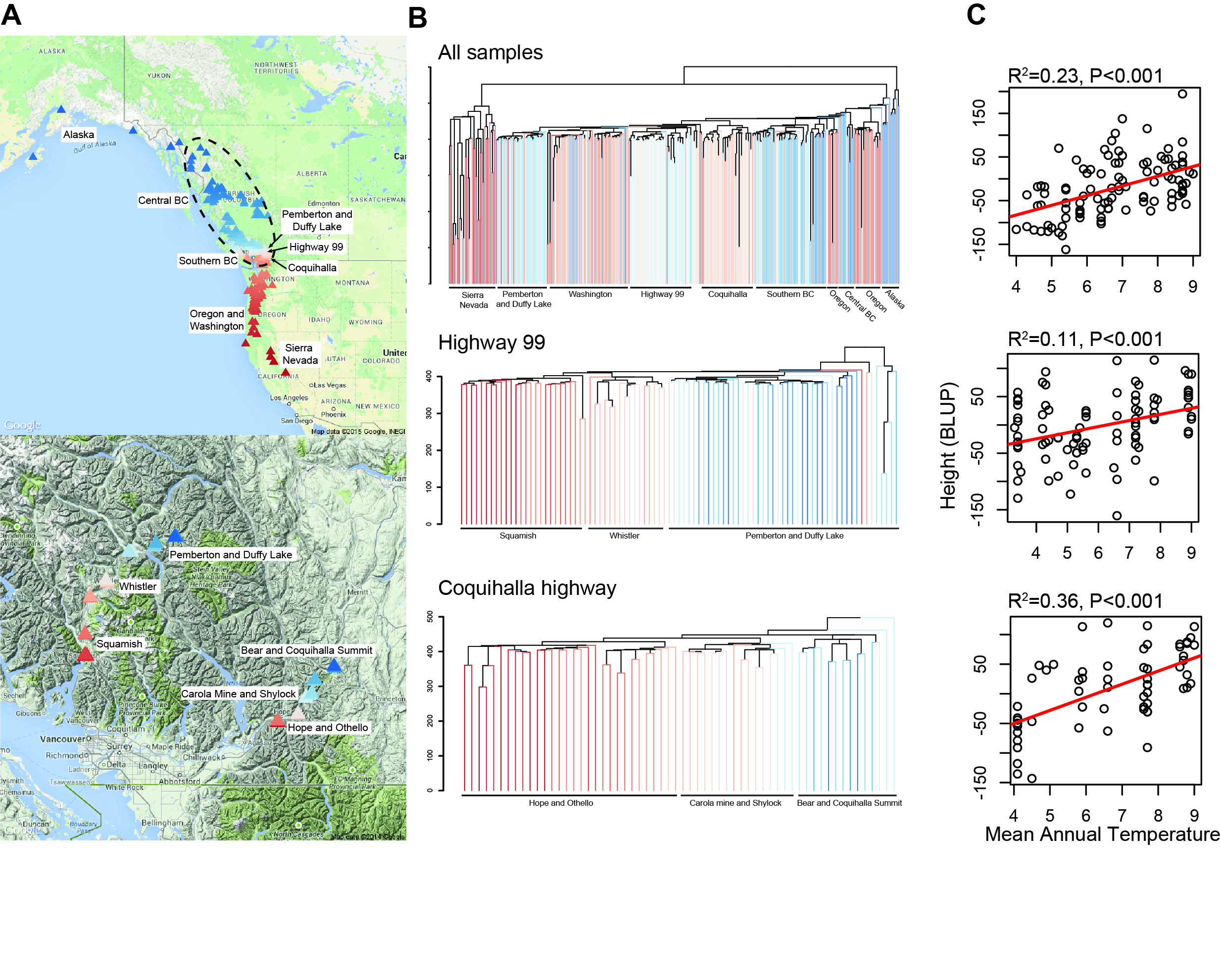
Poplar Hybridization
Due to their superior growth when compared to pure species, poplar hybrids comprise the vast majority of genotypes in operational plantations. However, our understanding of the genomic causes of hybrid vigor in outbred trees, and its consequences for local climatic adaptation, is limited. Natural Populus hybrid zones provide a ‘living laboratory’ in which there has been a long history of selection testing the genomic and phenotypic outcomes of hybridization. Through an NSF Plant Genome Research Project grant, we are combining extensive short and long-read genome resequencing for multiple latitudinal transects to exhaustively characterize genomic variation across these hybrid zones. We will then use admixture mapping to associate genomic ancestry with climate gradients and adaptive phenotypes, predict hybrid performance across a range of field test sites, and evaluate the contributions of environment and space in explaining patterns of hybridization. Finally, we will test the repeatability of hybrid outcomes using controlled crosses among Populus species, assess the predictive value of the loci identified as being involved in hybrid vigor/heterosis, and assess the role of allele-specific expression in generating hybrid phenotypes.